Amide Structure
The Unique Structure of Amide Bonds
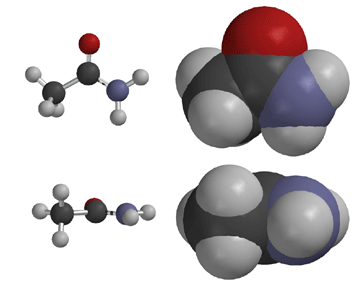
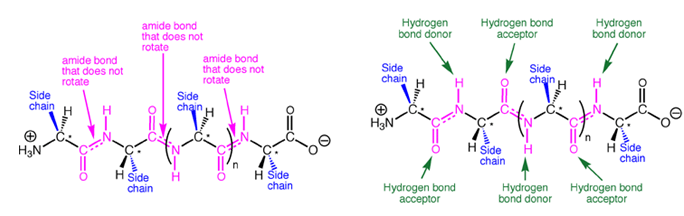
Amides have structural characteristics that are unique among carboxylic acid derivatives. Had you been asked in Chapter 1 to describe the geometry of an amide bond, you probably would have predicted bond angles of 120° about the carbonyl carbon and 109.5° about a tetrahedral amide nitrogen. In the late 1930s, one of Linus Pauling’s most important discoveries was that the geometry of the nitrogen atom of an amide bond in proteins is actually trigonal planar (sp2), not tetrahedral (sp3). A large number of subsequent experiments have verified that amides should be thought of as being best represented as the resonance hybrid of three resonance contributing structures, not two like other carbonyl species. In the unique third contributing structure of an amide, the lone pair on nitrogen donates electron density to create a pi bond between carbon and nitrogen.
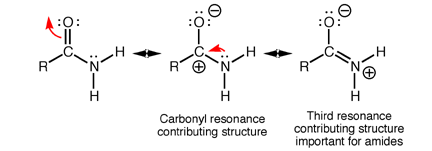
Inclusion of the third resonance contributing structure explains why the amide nitrogen is sp2 hybridized and therefore trigonal planar. Also, the presence of a partial double bond (pi bond) in the resonance hybrid indicates the presence of a restricted bond rotation about the C-N bond. In fact, the measured C-N bond rotation barrier is on the order of 63-84 kJ (15-20 kcal)/mol in amides, enough so that the bond does not rotate appreciably at room temperature. In addition, because the lone pair on nitrogen is delocalized into the pi bond, it is not as available for interacting with Lewis acids such as protons. Thus, amide nitrogens are not basic. In fact, in acid solution, amides are protonated on the carbonyl oxygen atom, not the nitrogen. Finally, the nitrogen lone pair electron delocalization also reduces the electrophilic character (partial positive charge) on the carbonyl carbon atom, reducing amide susceptibility to nucleophilic attack.

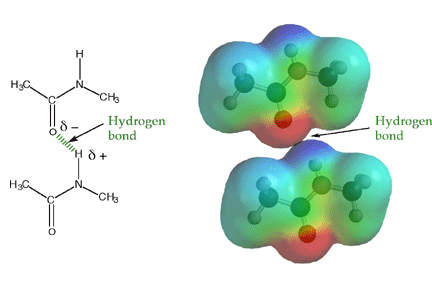
The amide -NH group is a good hydrogen bond donor, while the amide carbonyl is a good hydrogen bond acceptor, allowing primary and secondary amides to form strong hydrogen bonds.
Amide Bonds and Protein Folding
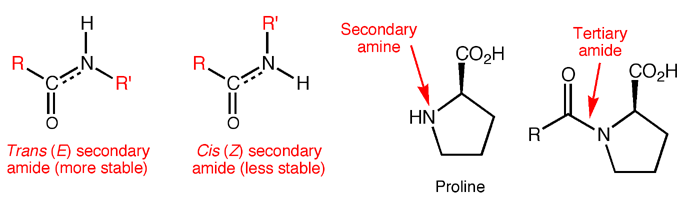
Proteins are made from amino acids connected through mostly secondary amide bonds, which adopt the more stable trans (E) configuration. This is thought to be due to repulsive steric interactions between side chains in the cis (Z) form that are not present in the trans (E) form, along with more favorable dipole-dipole interactions in the trans (E) form. Proline, the only amino acid to contain a secondary amine, produces tertiary amide bonds in proteins, so the cis and trans forms are of similar energy. In fact, proline cis-trans isomerism is now known to contribute substantially to the structure and function of certain proteins.
Because amide bonds are resistant to nucleophilic attack, the protein backbone is chemically very stable near physiological pH. Proteins are, however, susceptible to cleavage by certain specially adapted enzymes called proteases that can cleave amide bonds under mild conditions. Some proteases are the therapeutic targets of several recently introduced drugs.
Because of the large rotation barrier of the amide C-N bond, one out of every three bonds in a protein backbone does not rotate, significantly reducing the flexibility of the entire chain. In the context of protein folding, this conformational restriction is essential, because it requires much less stabilization energy to stably fold a chain with conformational restriction every third bond compared to a chain with no bond restrictions. The large number of hydrogen bonds possible along a protein backbone provide directional, non-covalent interactions that work in concert with other non-covalent interactions to stabilize a folded structure.
The folded three-dimensional structures of proteins endow them with the organization they need to carry out a remarkable array of catalytic activities that are the essence of the chemistry that keeps cells alive. The two basic patterns of protein folding are the alpha helix and beta sheet. A representative protein alpha helix and beta sheet are shown in the Figure with the trans amide bonds highlighted in magenta and backbone hydrogen bonds shown as green broken lines. The same backbone is shown above each structure for comparison, with side chains removed for clarity. The prominence of amide bonds in the protein backbone is apparent. These secondary structures are thus stabilized to a significant extent by the hydrogen bonding and conformational restriction of the protein backbone imposed by the amides.
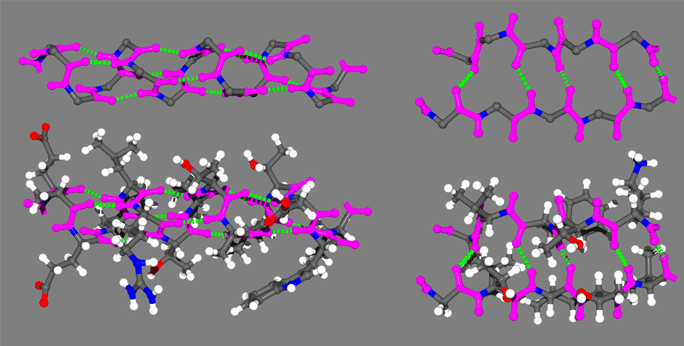
n the above figure are representative protein alpha helix (left) and beta strand (right). The four principle atoms of the backbone amide bonds are highlighted in magenta and backbone hydrogen bonds are shown as broken green lines. The upper structures are the backbone only, with side chain atoms removed for clarity.
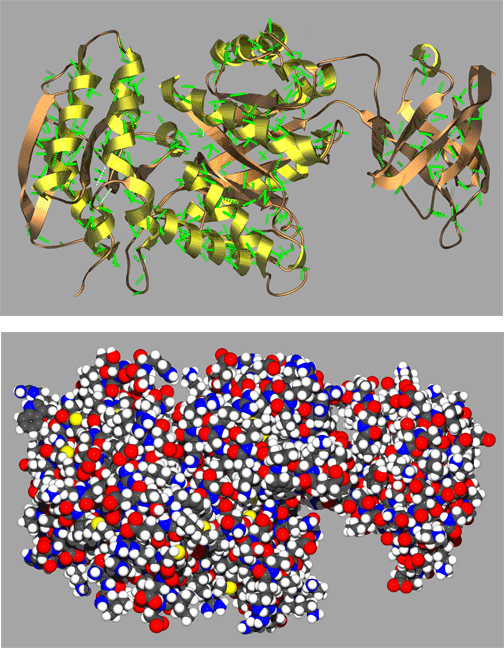
A representative complete protein structures in Figure Y illustrates just a small sample of the remarkably complex and often quite beautiful folding patterns found in proteins. Here the protein is shown in both a spacefilling and ribbon cartoon model with coils representing alpha helices and the arrows representing strands of a beta sheet. Scientists often use visual aids such as these ribbon cartoons to help them visualize complex structures. Proetin structures such as these would not be stable if it were not for the hydrogen bonding and conformational restriction imparted by amide bonds of the protein backbone. It is reasonable to say that the unique structural characteristics of amide bonds have played a relatively large part in shaping the evolution of life on our planet!




Pictures of the Day
2-18-2025